This article also appears in Canadian Developer Connection.

Intro
Here’s the second of my notes from the Science 2.0 conference, a conference for scientists who want to know how software and the web is changing the way they work. It was held on the afternoon of Wednesday, July 29th at the MaRS Centre in downtown Toronto and attended by 102 people. It was a little different from most of the conferences I attend, where the primary focus is on writing software for its own sake; this one was about writing or using software in the course of doing scientific work.
My previous notes from the conference:
This entry contains my notes from Cameron Neylon’s presentation, A Web Native Research Record – Applying the Best of the Web to the Lab Notebook.
Here’s the abstract:
Best practice in software development can save researchers time and energy in the critical analysis of data but the same principles can also be applied more generally to recording research process. Successful design patterns on the web tend to be those that successfully couple people into efficient information transfer mechanisms. Can we re-think the way we create, keep, and share our research records by using these design patterns to make it more effective?
Here’s Cameron’s bio:
Cameron Neylon is a biophysicist who has always worked in interdisciplinary areas and is a leading advocate of data availability. He currently works as Senior Scientist in Biomolecular Sciences at the ISIS Neutron Scattering facility at the Science and Technology Facilities Council. He writes and speaks regularly on the interface of web technology with science and is well-known as one of the leading proponents of open science.
The Notes
- Feel free to copy and remix this presentation – it’s licenced under Creative Commons
- What is the web good for?
- Publishing
- Subscribing
- Syndicate
- Remix, mash up and generally do stuff with
- Collaborate
- What do scientists do?
- Publish
- Syndicate (CRC books are a form of syndication)
- Remix (take stuff from different disciplines — pull things to toghter, remix them
- Validate
- Collaborate
- So, with this overlap, the web has solved science problems, right?
- No — papers are dead, broken and disconnected
- Papers don’t have links
- The whole scientific record is fundamentally a dead document
- The links between things make the web go round
- I want to make science less like a great big monolithic document and make it more like a network of pieces of knowledge, wired together:
- Fragments of science
- Loosely coupled
- Tightly wired
- No — papers are dead, broken and disconnected

- What is a “fragment of science”?
- A paper is too big a piece, even if it is the "minimal publishable unit"
- A tweet is too small
- A blog post would be the right size
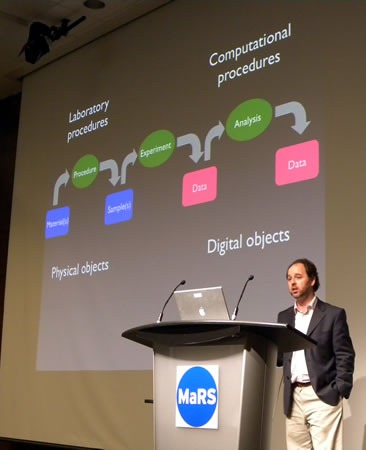
- His lab book is a collection of various electronic documents:
- Excel files
- Some basic version control
- Data linked back to description of process used to create the data
- As far as possible, the blogging is done automatically by machines
- It doesn’t have to be complicated
- [Shows a scatter plot, with each point representing an experiment]:
- Can we tell an experiment didn’t work by its position on the graph?
- We can tell which experiments weren’t recorded properly – they have no links to other experiments
- The use of tagging and “folksonomies” goes some way, but how do you enforce it?
- Tags are Inconsistent — not just between people, but even within a single person – you might tag the same thing differently from day to day
- Templates create a virtuous circle, a self-assembling ontology
- We found that in tagging, people were mixing up process and characteristics – this tells us something about the ontology process
- Put your data in external services where appropriate
- Flickr for images
- YouTube for video
- RCSBPDB Protein Data Bank
- Chemspider
- Even Second Life can be used as a graphing medium!
- All these services know how to deal with specific data types
- Samples can be offloaded
- LIMS, database, blogs, wiki, spreadsheet
- Procedures are just documents
- Reuse existing services
- Semantic feed of relationships — harness Google: most used is the top result
- Semantic web creates UI issues
- Just trying to add meaning to results is one step beyond what scientists are expected to do
- We need a collaborative document environment
- The document environment must feel natural for people to work in
- When they type something relevant, the system should realize that and automatically link it
- We’re at the point where doc authoring systems can use regular expressions to recognize relevant words and autolink them

- The current mainstream response to these ideas is:
- The gamut from "You mean facebook?" to horror
- I’m not worried about these ideas not getting adopted
- Scientists are driven by impact and recognition
- How do we measure impact?
- Right now, we do this by counting the number of papers for which you’re an author
- Most of my output is not published in traditional literature; it’s published freely on the web for other people to use
- If they’re not on the web, they disappear from the net
- The future measure of your scientific impact will be its effect on the global body of knowledge
- Competition will drive adoption
- How do we measure impact?