This article also appears in Canadian Developer Connection.

Here’s the third in a series of notes from the Science 2.0 conference, a conference for scientists who want to know how software and the web is changing the way they work. It was held on the afternoon of Wednesday, July 29th at the MaRS Centre in downtown Toronto and attended by 102 people. It was a little different from most of the conferences I attend, where the primary focus is on writing software for its own sake; this one was about writing or using software in the course of doing scientific work.
My previous notes from the conference:
- Choosing Infrastructure and Testing Tools for Scientific Software Projects – C. Titus Brown
- A Web Native Research Record – Applying the Best of the Web to the Lab Notebook – Cameron Neylon
This entry contains my notes from Victoria Stodden’s presentation, How Computational Science is Changing the Scientific Method.
Here’s the abstract:
As computation becomes more pervasive in scientific research, it seems to have become a mode of discovery in itself, a “third branch” of the scientific method. Greater computation also facilitates transparency in research through the unprecedented ease of communication of the associated code and data, but typically code and data are not made available and we are missing a crucial opportunity to control for error, the central motivation of the scientific method, through reproducibility. In this talk I explore these two changes to the scientific method and present possible ways to bring reproducibility into today’ scientific endeavor. I propose a licensing structure for all components of the research, called the “Reproducible Research Standard”, to align intellectual property law with longstanding communitarian scientific norms and encourage greater error control and verifiability in computational science.
Here’s her bio:
Victoria Stodden is the Law and Innovation Fellow at the Internet and Society Project at Yale Law School, and a Fellow at Science Commons. She was previously a Fellow at Harvard’s Berkman Center and postdoctoral fellow with the Innovation and Entrepreneurship Group at the MIT Sloan School of Management. She obtained a PhD in Statistics from Stanford University, and an MLS from Stanford Law School.
The Notes
- Research has been how massive computation has changed the practice of science and the scientific method
- Do we have new modes of knowledge discovery?
- Are standards of what we considered knowledge changing?
- Why aren’t researchers sharing?
- One of my concerns is facilitating reproducibility
- The Reproducible Research Standard
- Tools for attribution and research transmission
- Example: Community Climate Model
- Collaborative system simulation
- There are community models available
- Built on open code, data
- If you want to model something a complex as climate, you need data from different fields
- Hence, it’s open
- Example: High energy physics
- Enormous data produced at LHC at CERN — 15 petabytes annually
- Data shared through grid
- CERN director: 10 – 20 years ago, we might have been able to repeat an experiment – they were cheaper, simpler and on a smaller scale. Today, that’s not the case
- Example: Astrophysics
- Data and code sharing, even among amateurs uploading their photos
- Simulations: This isn’t new: even in the mid-1930s, they were trying to calculate the motion of cosmic rays in Earth’s magnetic field via simulation
- Example: Proofs
- Mathematical proof via simulation vs deduction
- My thesis was proof via simulation – the results were not controversial, but the methodology was

- The rise of a “Third Branch” of the Scientific Method
- Branch 1: Deductive/Theory: math, logic
- Branch 2: Inductive/Empirical: the machinery of hypothesis testing – statistical analysis of controlled experiments
- Branch 3: Large-scale extrapolation and prediction – are we gaining knowledge from computation/simulations, or they just tools for inductive reasoning?
- Contention — is it a 3rd branch?
- See Chris Anderson’s article, The End of Theory (Wired, June 2008)
- Systems that explain the world without a theoretical underpinning?
- There’s the “Hillis rebuttal”: Even with simulations, we’re looking for patterns first, then create hypotheses, the way we always have
- Steve Weinstein’s idea: Simulation underlies both branches:
- It’s a tool to build intuition
- It’s also a tool to test hypotheses
- Simulations let us manipulate systems you can’t fit in a lab
- Controlling error is central to scientific process

- Computation is increasingly pervasive in science
- In the Journal of the American Statistical Association (JASA):
- In 1996: 9 out of 20 articles published were computational
- In 2006: 33 out 35 articles published were computational
- In the Journal of the American Statistical Association (JASA):
- There’s an emerging credibility crisis in computational science
- Error control forgotten? Typical scientific computation papers don’t include code and data
- Published computational science is near impossible to replicate
- JASA June 1996: None of the computational papers provided any code
- JASA June 2006: Only 3 out of the 33 computational articles made their code publicly available
- Changes in scientific computation:
- Internet: Communication of all computational research details and data is possible
- Scientists often post papers but not their complete body of research
- Changes coming: Madagascar, Sweave, individual efforts, journal requirements
- A potential solution: Really reproducible research
- The idea of an article as not being the scholarship, but merely the advertisement of that scholarship
- Reproducibility: can a member of the field independently verify the result?

- Barriers to sharing
- Took a survey of computational scientists
- My hypotheses, based on the literature of scientific sociology:
- Scientists are primarily motivated by personal gain or loss
- Scientists are primarily worried about being “scooped”
- Survey:
- The people I surveyed were from the same subfield: Machine learning
- They were American academics registered at a top machine learning conference (NIPS)
- Respondents: 134 responses from 638 requests (23%, impressive)
- They were all from the same legal environment of American intellectual property
- Based on comments, it’s in the back of people’s minds
- Reported sharing habits
- 32% put their code available on the web
- 48% put their data
- 81% claimed to reveal their code
- 84% said their data was revealed
- Visual inspection of their sites revealed:
- 30% had some code posted
- 20% had some data posted
- Reported sharing habits
- Preliminary findings:
- Surprising: They were motivated to share by communitarian ideals
- Surprising: They were concerned about copyright issues
- Barriers to sharing: legal
- The original expression of ideas falls under copyright by default
- Copyright creates exclusive right of author to:
- Reproduce work
- Prepare derivative works
- Creative Commons
- Make it easier for artists to share and use creative works
- A suite of licences that allows the author to determine the terms
- Licences:
- BY (attribution)
- NC (non-commercial)
- ND (no derived work)
- SA (share-alike)
- Open Source Software Licencing
- Creative Commons follows the licencing approach used for open source software, but adapted for creative works
- Code licences:
- BSD licence: attribution
- GPL: attribution and share-alike
- Can this be applied to scientific work?
- The goal is to remove copyright’s block to fully reproducible research
- Attach a licence with an attribution to all elements of the research compendium

- Proposal: Reproducible research standard
- Release media components (text, data) under CC BY
- Code: Modified BSD or MIT (attrib only)
- Releasing data
- Raw facts alone are generally not copyrightable
- Selection or arrangement of data results in a protected compilation only if the end result is an original intellectual creation (US and Canada)
- Subsequently qualified: facts not copied from another source can be subject to copyright protection
- Benefits of RRS
- Changes the discussion from "here’s my paper and results" to "here’s my compendium”
- "Gives funders, journals and universities a “hook”
- If your funding is public, so should your work!
- Standardization avoids licence incompatibiltiies
- Clarity of rights beyond fair use
- IP framework that supports scientific norms
- Facilitation of research, thus citation and discovery
- Reproducibility is Subtle
- Simple case: Open data and small scripts. Suits simple definition
- Hard case: Inscrutable code; organic programming
- Harder case: Massive computing platforms, streaming sensor data
- Can we have reproducibility in the hard cases?
- Where are acceptable limits on non-reproducibility?
- Privacy
- Experimental deisgn
- Solutions for harder cases
- Tools
- Openness and Taleb’s criticism
- Scientists are worried about contamination by amateurs
- Also concerned about the “Prisoner’s dilemma”: they’re happy to share their work, but not until everyone else does


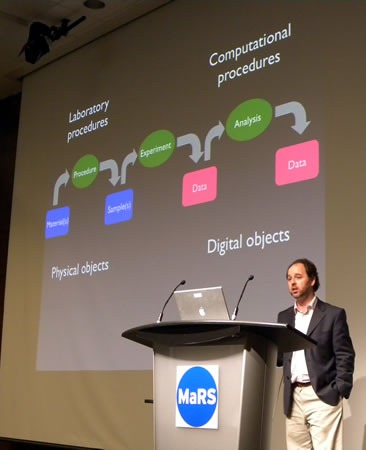

 C. Titus Brown delivering his presentation.
C. Titus Brown delivering his presentation.
